Markus Covert Lab
Causality Network
The Markus Covert Lab at Stanford built an unprecedented whole-cell model. Realizing their existing process for exploring and updating the model would only find or confirm what they already knew, the Covert team approached us about finding a more accessible and insightful way to work with their information.
Capabilities
- Custom tool design, including conceptualization, design, back-end and front-end development
Challenges
- How can we make working with 30,000+ nodes and measurements feel smooth and manageable?
- The lab needed a way to facilitate serendipitious discovery, model debugging, and simulation comparison. How can we create one tool for all of those tasks?
Outcomes
- A completely custom tool, tailored to the Covert Lab’s workflow
- Replaced binders of printouts, manual searching, and siloed Python scripts with a central application that allowed everyone to be on the same page, but also complete their individual tasks
The model allows researchers to run simulations of a single cell. It aggregates decades of research (the team combed through ten thousand papers, millions of data points, and hundreds of labs) and provides huge possibilities for biological discovery and medical research. When the model was first completed, identifying bugs and verifying hypotheses, meant keyword searches with enormous text files or writing custom Python code to make sense of the outputs. Realizing that this process would only find or confirm what they already knew, the Covert team approached us about finding a more accessible and insightful way to work with their information.
There are 30,000 molecules and processes included in the model, each with thousands of measurements. Within the cacophony of these outputs, the team needed to be able to test and discover new causal relationships to further their research. Through a series of collaborations, we worked with the Covert Lab to build meaningful representations and interactions that matched how they wanted to think about their data.
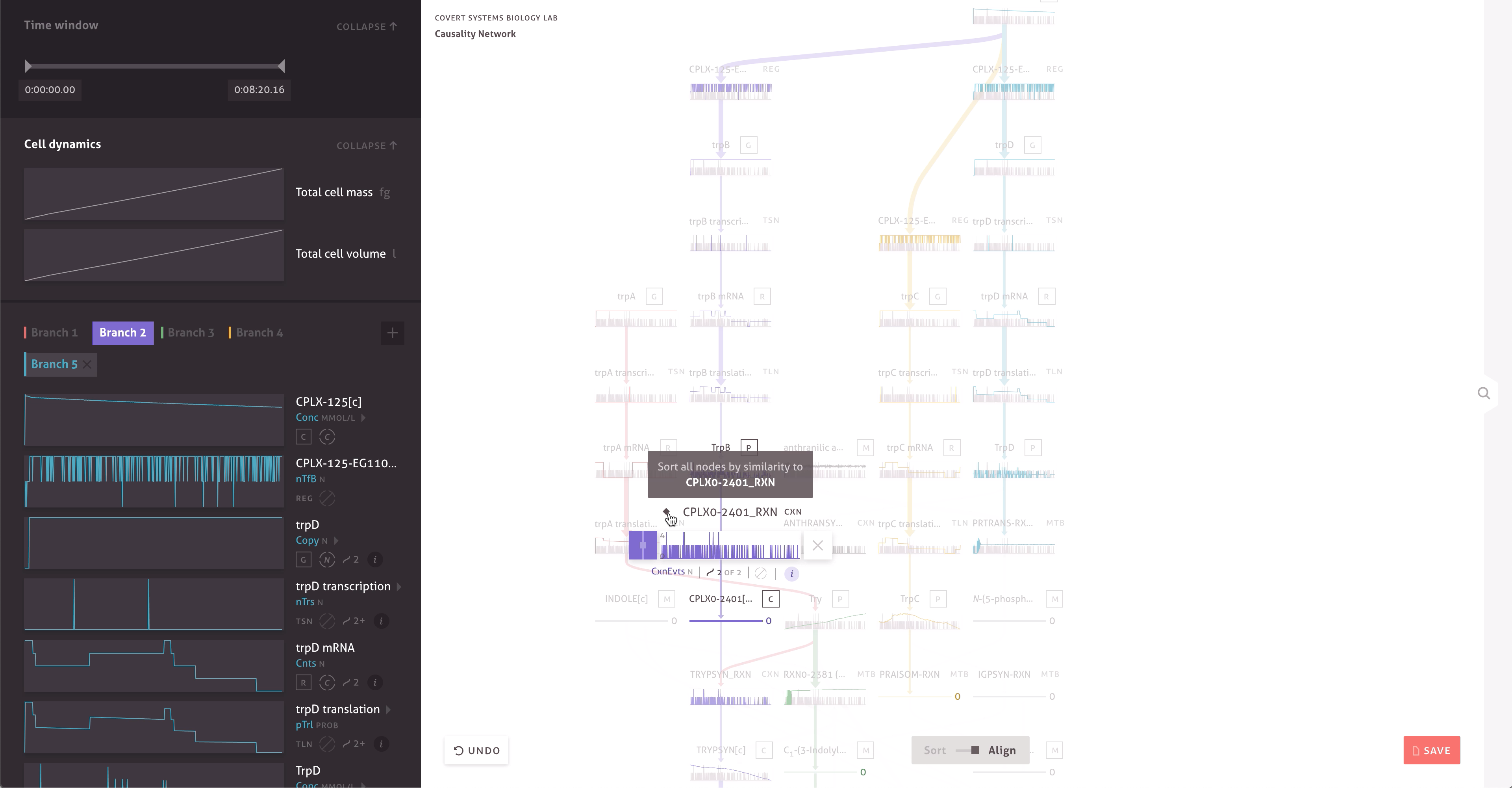
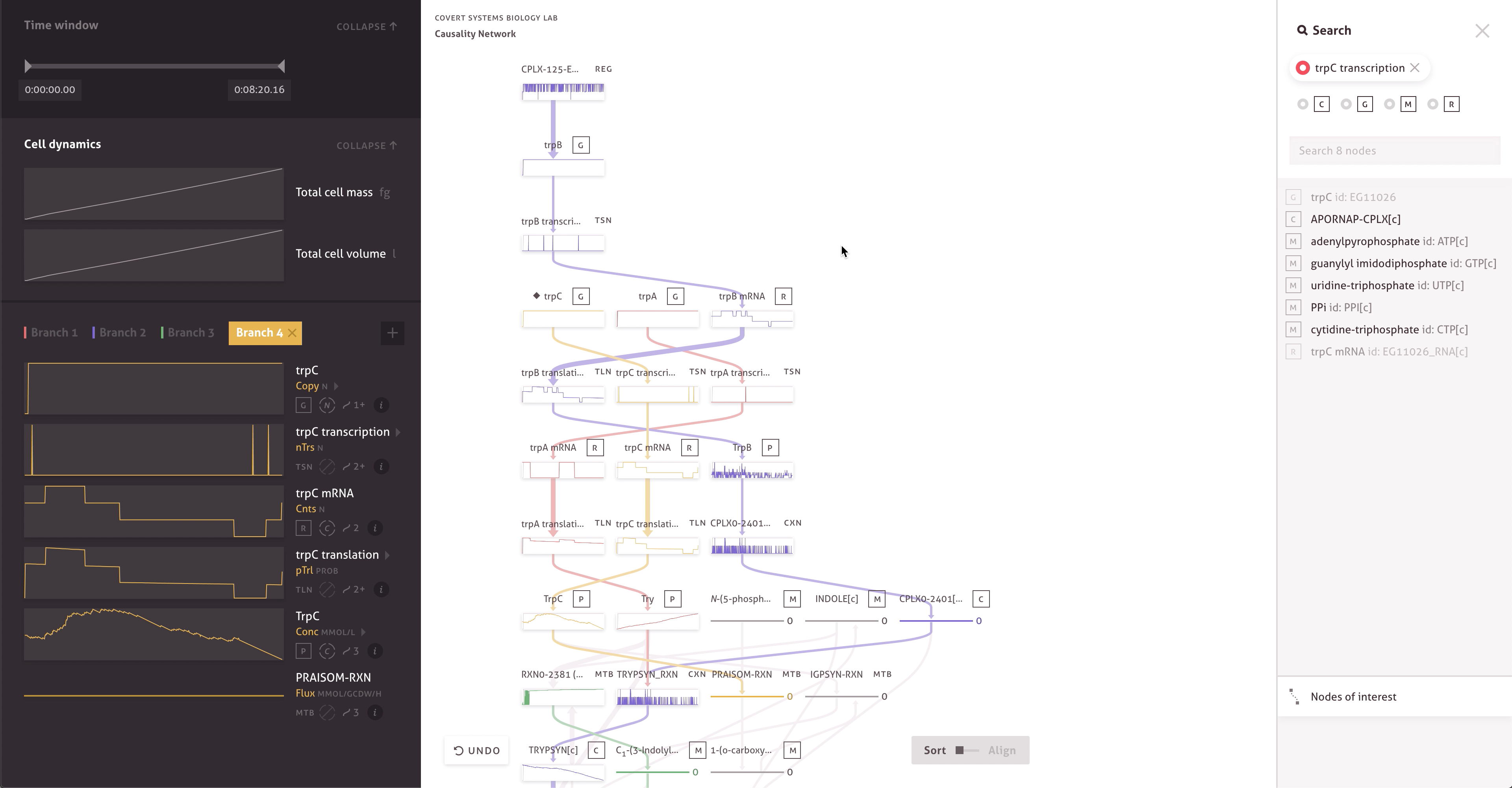
The final result far surpasses simple text searches and custom scripts, providing the team with ways to easily find serendipitous connections, construct causal pathways, and compare the outputs of multiple simulations. It allows team members to get a more intuitive sense of the data by enabling quick hypothesis testing and rapid iteration. When responding to peer reviewers or following up on research, the team can even turn to the application and quickly check simulation outputs.