The Sabeti Lab | Sentinel
Lookout
With the support of the Sabeti Lab, we have built Lookout: a tool for pathogen detection, tracking, and response.
Capabilities
- Rapid prototyping in a fast-paced, changing situation
- Scalable deployment for tens-of-thousands of users
- Front-end design and development
- Building intuitive tools for complex, real-time information

Challenges
- How can we enable administrators and staff with little to no experience in pandemic response to become experts?
- How can we bring all data sources together to facilitate critical decision-making in real-time?
Achievements
- Created a tool that brings all relevant pathogen tracking and contact tracing data into one place.
- Supported a college campus of 10,000+ students in their commitment to bringing students back in person
In its original instantiation, Lookout paired COVID-19 specific data (ex: testing, viral sequencing, wastewater) with institution information (ex: work and recreational teams, floorplans, schedules, etc.) to empower any sized health and safety or contact tracing team to better understand and make decisions around their data.
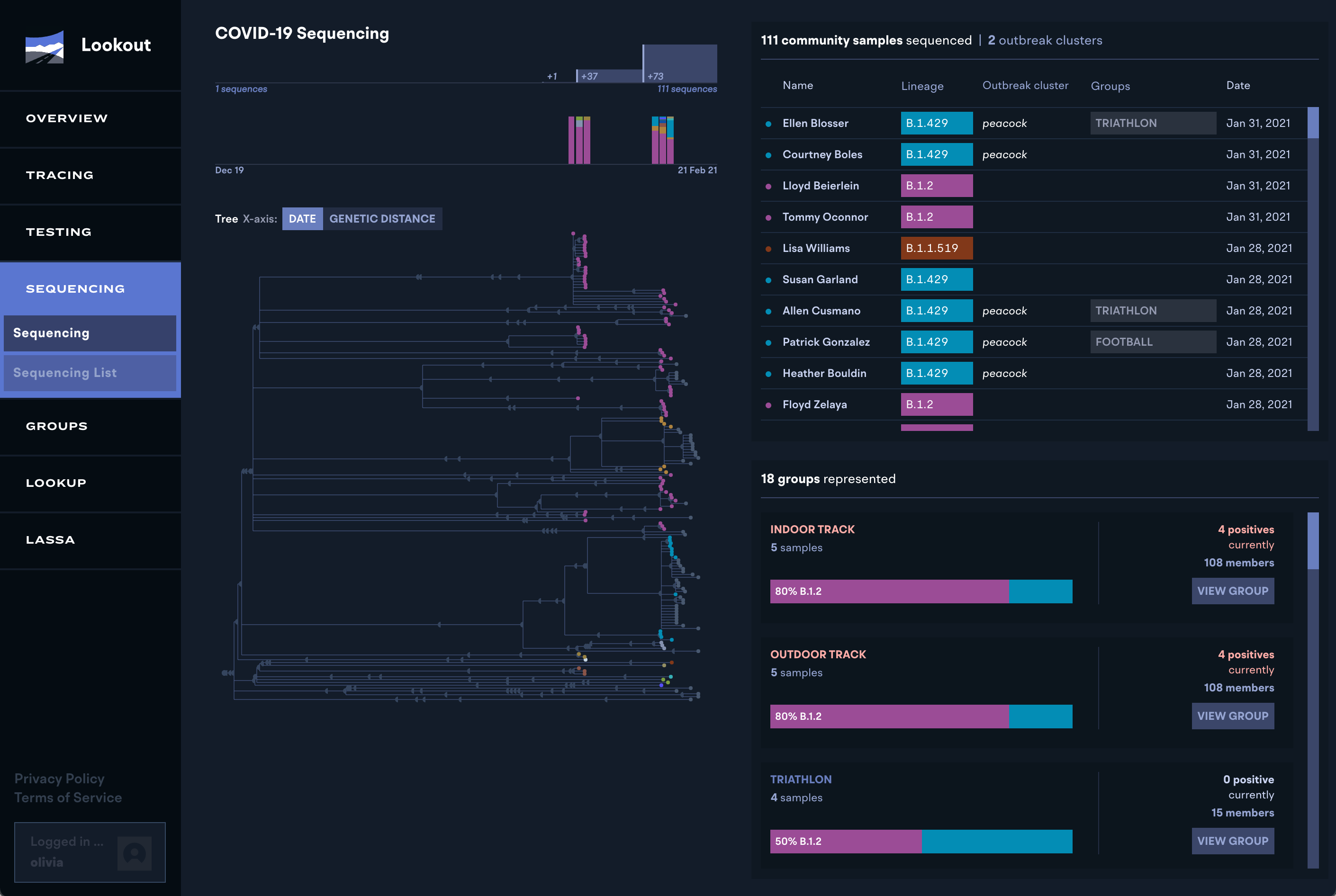
Part of our core focus from the beginning was finding ways to integrate traditional contact tracing and response information with cutting edge viral genomic and multiplex testing information. In our deployment at Colorado Mesa University, response staff could see which lineages were spreading on campus, as well as how many cases were inter-community spread versus new introductions and tailor their response based on that information if needed.
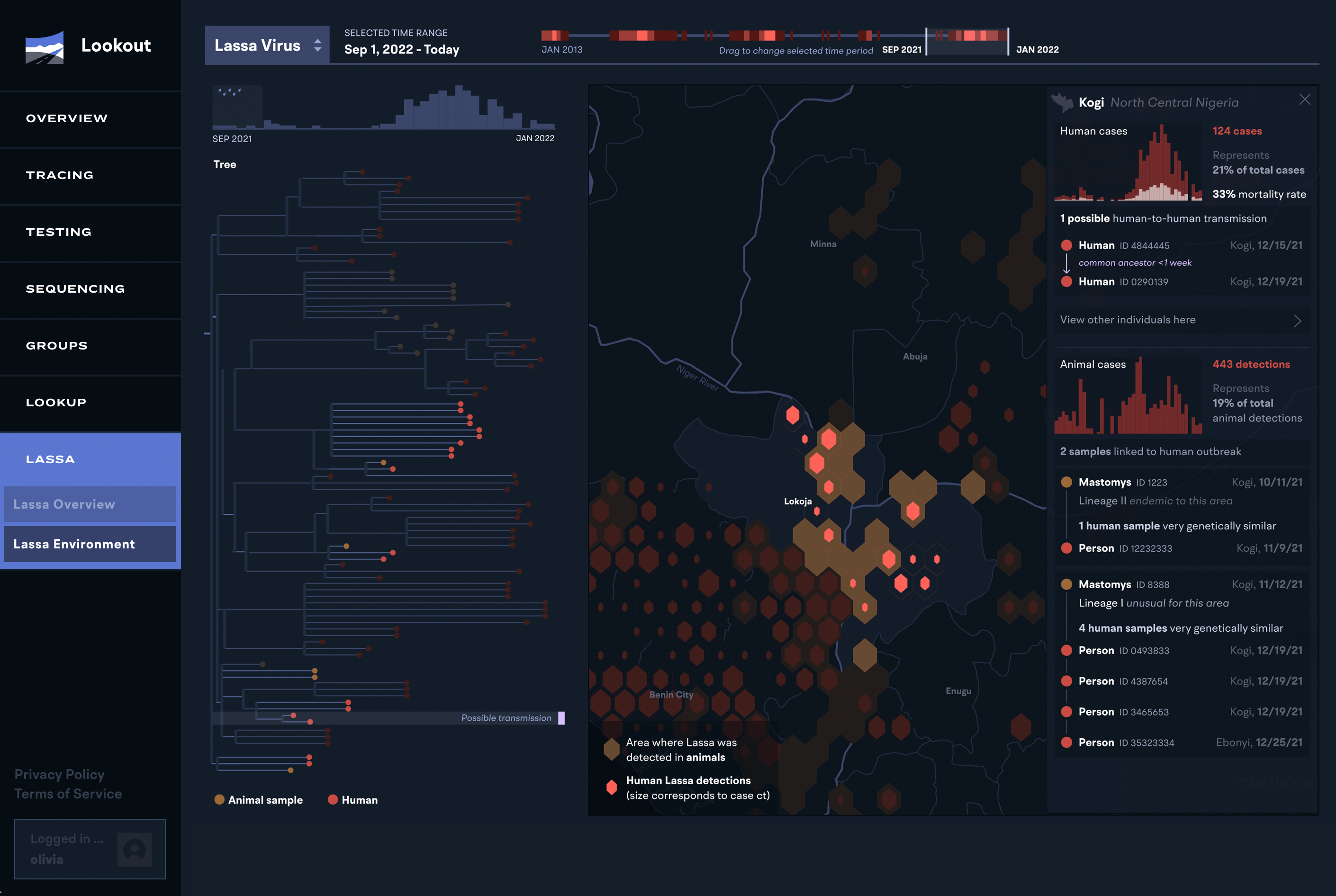
In continuing our development on Lookout, we have expanded it to track the spread and contact tracing information for any pathogen. As we’ve moved into working with multiple diseases, we’ve worked on both data standardization, as well as continuing to build out pathogen-specific views tailored to the different types of tracking and response. We’ve built a robust API for integrating and updating data into the system.
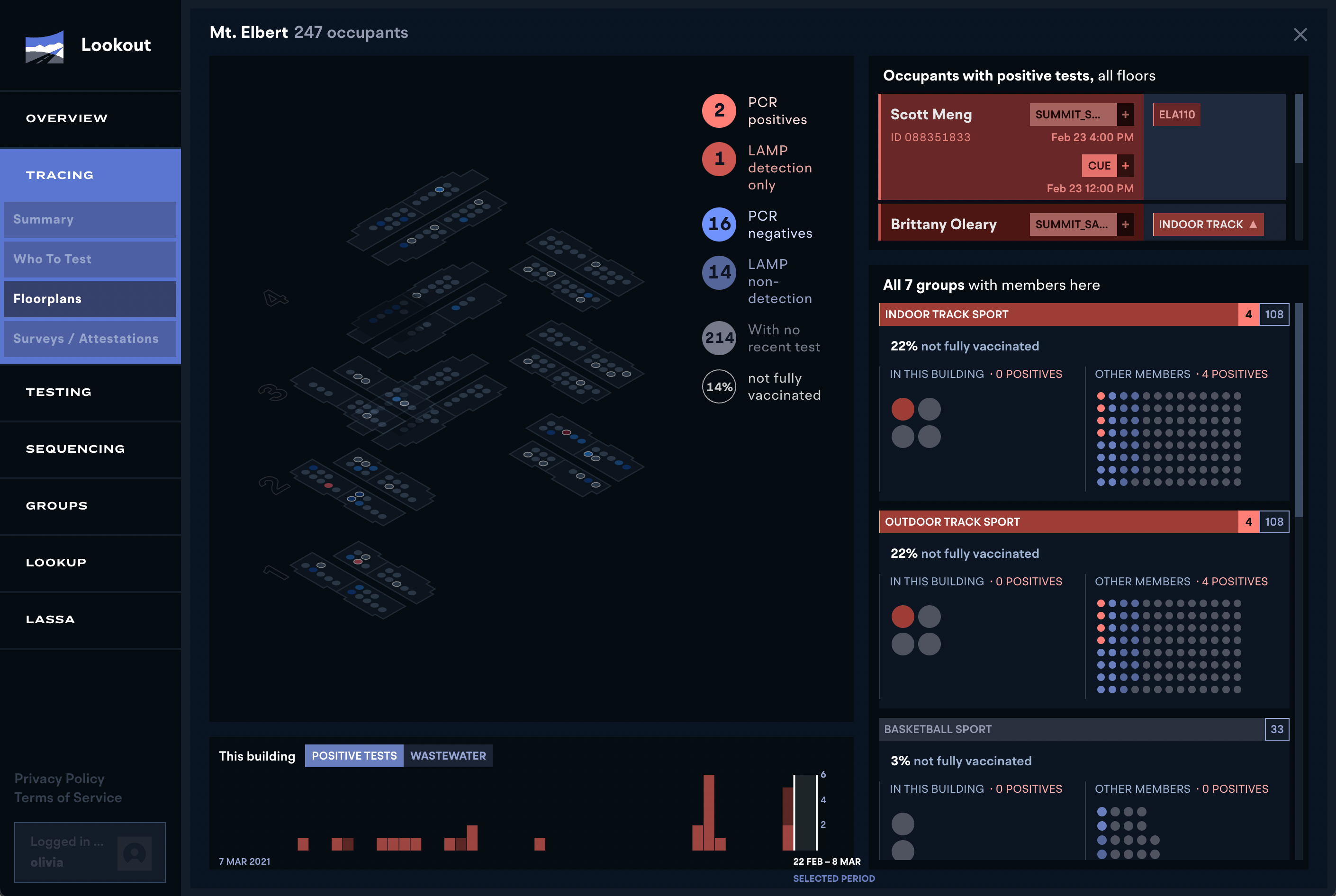
More about Lookout's role in the COVID-19 response at Colorado Mesa University can be found here.